The Power of Value_Locate
QUESTION: I've read a couple of articles on your web page that utilize the IDL command Value_Locate in unusual and impressive ways. It is getting to be one of my favorite commands, taking its place right up there with Histogram and Where for their power and usefulness. It is hard to get a sense of what the command actually does from the IDL documentation. Can you give a few examples that illustrate its power?
![]()
ANSWER: Jeremy Bailin suprised me the other day with an extremely clever way of partitioning array values into discrete colors for display with the Value_Locate command. I asked him if he would like to write a guest tutorial that would unlock the power of Value_Locate for the rest of us. Here is what he came up with.
![]()
There are probably two reasons why Value_Locate is underused. The first is that it was only introduced in IDL 5.3, well after many people developed their core techniques. The second is that the help page is somewhat opaque on what it actually does. The basic idea is pretty simple: given two arrays values and array,
result = Value_Locate(values, array)
tells you where within values the elements of array are located. A concrete example will help:
IDL> values = [-10, -5, 23, 109]
IDL> arrray = [-5, -5, 23, 23, -10, 109]
IDL> Print, Value_Locate(values, array) 1 1 2 2 0 3
IDL> Print, values[Value_Locate(values, array)]
-5 -5 23 23 -10 109
In other words, -5 is element number 1 of values, 23 is element number 2, -10 is element number 0, and 109 is element number 3. They are the subscripts of the located elements within values. And, indeed, if we subscript values by those indices, we end up with the original array.
One important caveat is that values must be strictly increasing. You will get nonsense answers otherwise.
IDL> values = [-10, 23, 109, -5] IDL> arrray = [-5, -5, 23, 23, -10, 109]
IDL> Print, Value_Locate(values, arrray) 0 0 1 1 0 3
IDL> Print, values[Value_Locate(values, arrray)]
-10 -10 23 23 -10 -5
Technically, values can also be monotonically decreasing, but the return value doesn't have exactly the same meaning. I recommend sticking with the monotonically increasing case.
![]()
Mapping Between Sets
I use Value_Locate in this vein all the time as a way of creating a mapping between the set of integers and any other finite set of numbers. Let's say I have an array whose elements can only take on the following five values: -23.5, 19.4, 2.0, -9999, 14.1. For a great many purposes (such as the Histogram examples that I'll get to below) integers between 0 and 4 are a much nicer set of numbers to deal with than this smorgasbord of floating point numbers. We can map back and forth between these two representations quite easily using Value_Locate.
IDL> arrray = [2.0, 2.0, 19.4, -9999, 14.1, -9999, 19.4, -9999, 2.0]
IDL> values = [-23.5, 19.4, 2.0, -9999, 14.1]
IDL> values = values[Sort(values)]
IDL> mappedArray = Value_Locate(values, arrray) IDL> print, mappedArray
2 2 4 0 3 0 4 0 2
Here we have taken some floating point data and converted it into a
much simpler set of integers. Note that we had to sort values first with
the Sort command to make sure the elements in values were in a monotonically increasing
order. If we have an array of integers, the reverse operations is equally simple.
IDL> mappedResult = [1, 1, 3, 4, 0, 0]
IDL> Print, values[mappedResult]
-23.5000 -23.5000 14.1000 19.4000 -9999.00 -9999.00
This is in some ways similar to the enumeration type that is available in C and some other languages.
We can also map between two different non-integer enumerations by sticking a forward mapping from one onto the reverse mapping of the other.
IDL> values1 = [-9999, -23.5, 2.0, 14.1, 19.4]
IDL> values2 = [ 100, 100.5, 101, 101.5, 102]
IDL> arrray = [14.1, -23.5, -9999, 2.0]
IDL> Print, values2[Value_Locate(values1, arrray)]
101.500 100.500 100.000 101.000
Here -9999 gets mapped to 100, -23.5 gets mapped to 100.5, etc.
![]()
Ranges
In every example so far, each element of array occurs exactly within values. But what if some of the elements don't appear there? Let's try it out!
IDL> values = [0, 10, 20, 30]
IDL> array = [-5, 5, 15, 25, 35] IDL> Print, Value_Locate(values, array)
-1 0 1 2 3
Here is a table of what we have so far.
Values: 0 10 20 30 Index: 0 1 2 3 Array: -5 5 15 25 35 Return: -1 0 1 2 3
We see that if an element of array lies between two elements of values, Value_Locate rounds down to the lower index. For example, 15 lies between 10 (index 1) and 20 (index 2), so Value_Locate returns 1. This rounding down even occurs when values of array lie outside of the range of values: in the case above, -5 is less than values[0], so Value_Locate rounds down to -1; similarly, 35 rounds down to the highest value, values[3], and so Value_Locate returns 3.
Using Ranges For Partitioning
There are many applications for using Value_Locate in this manner. One example is partitioning floating point data into unevenly-spaced bins for display purposes. To repeat the example from the Array Partitioning article, say you have a 2D array of values ranging from 0 to 1, and want to display it as an image with a small number of colors, depending on the value in the array.
< 0.2: white 0.2 - 0.3: green 0.3 - 0.5: yellow 0.5 - 0.8: blue > 0.8: red
The first thing to do is to set up a color table that loads white into color index 1, green into 2, etc. But how do we then turn our floating point values into color indices? With Value_Locate, it's simple.
IDL> data = cgDemoData(11) IDL> array = cgScaleVector(array, 0.0, 1.0) IDL> cutoffs = [0.2, 0.3, 0.5, 0.8] IDL> image = Byte(Value_Locate(cutoffs, array) + 2)
What have we done here? We have asked where each floating point number in array would fall in cutoffs. All of the ones that lie below 0.2 will return -1, all of the ones that lie between 0.2 and 0.3 will return 0, all of the ones that lie between 0.3 and 0.5 will return 1, etc. We just need to add 2 to get to the color index for each range, and convert to byte type for displaying.
You can see an example in the figure below of what has happened. On the left side of the figure is a histogram of the original data. On the right side of the figure is a histogram of the partitioned data. Note that it has been partitioned into just five values. Here is the code that produced the figure.
IDL> Window, XSIZE=700, YSIZE=400, TITLE='Value_Locate Used to Partition Data' IDL> !P.Multi = [0,2,1] IDL> cgHistoplot, array, BINSIZE=0.025, /FILL IDL> cgHistoplot, image, BINSIZE=1.0, /FILL IDL> !P.Multi = 0
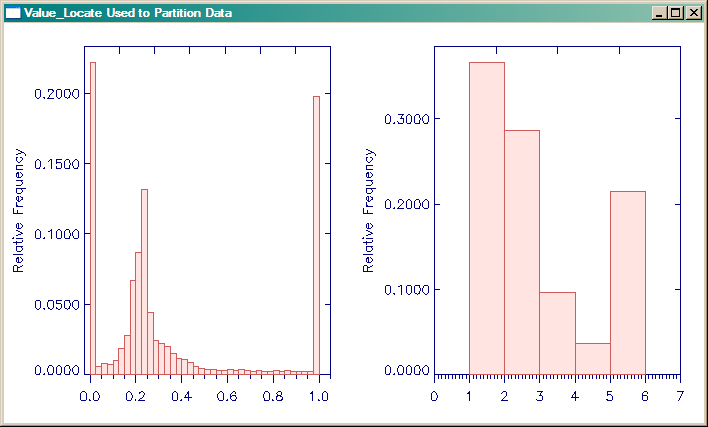 |
| Value_Locate used to partition data into discrete color values. |
Using Ranges in an Interpolation Scheme
Another convenient use of this property of Value_Locate is for interpolation. Most interpolation schemes work by fitting a low-order polynomial or similar function to the points near the desired location. How do you efficiently determine which points are the "near points?" Using Value_Locate!
The simplest example is a linear interpolation between the neighbouring points: if x[i] &le array[j] &le x[i+1] (where x is strictly increasing), then the interpolated value is:
interpolated_y[j] = y[i] * (x[i+1]-array[j])/(x[i+1]-x[i]) + y[i+1] * (array[j]-x[i])/(x[i+1]-x[i])
The trick is to figure out which i to use for a given j. But that's exactly what Value_Locate does. Here is some simple code that will calculate this interpolation (I haven't taken care to handle the edge cases correctly, but see the code of the library function Interpol for more details):
IDL> left = Value_Locate(x, array)
IDL> right = left+1
IDL> interpolated_y = y[left] * (x[right]-array)/(x[right]-x[left]) + y[right] $
* (array-x[left])/(x[right-x[left])
This is equivalent to Interpol(y, x, array).
![]()
A Serving of Value_Locate With A Side Of Histogram
Histogram has a well-deserved reputation as the foundation of most IDL optimization strategies because of its combination of speed and the wonderful Reverse_Indices facility. However, some problems appear difficult to solve using Histogram because it can only use fixed bin sizes. As I'll demonstrate below, Value_Locate can be coupled with Histogram to make it even more powerful (yikes!).
A common question to answer with Histogram is "which elements lie in each bin?" This is straightforward if we have equally spaced bins, but what if we want our bin edges to be spaced non-uniformly?
The trick is to get Value_Locate to partition the data into integers, and then run Histogram on the uniformly-spaced integers that result. For example, consider this code.
IDL> cutoffs = [0.2, 0.3, 0.5, 0.8] IDL> data = RandomU(43L, 10)
IDL> Print, data
0.331022 0.151196 0.114072 0.203458 0.0409741 0.614608
0.951897 0.191795 0.0152987 0.709563
IDL> mappedData = Value_Locate(cutoffs, data) IDL> Print, mappedData
1 -1 -1 0 -1 2 3 -1 -1 2
IDL> h = Histogram(mappedData, MIN=-1, REVERSE_INDICES=ri) IDL> Print, h
5 1 1 2 1
IDL> Print, data[ri[ri[0]:ri[1]-1]] ; Values less than 0.2.
0.151196 0.114072 0.0409741 0.191795 0.0152987
IDL> Print, data[ri[ri[1]:ri[2]-1]] ; Values between 0.2 and 0.3.
0.203458
IDL> Print, data[ri[ri[2]:ri[3]-1]] ; Values between 0.3 and 0.5.
0.331022
IDL> Print, data[ri[ri[3]:ri[4]-1]] ; Values between 0.5 and 0.8.
0.614608 0.709563
IDL> Print, data[ri[ri[4]:ri[5]-1]] ; Values greater than 0.8.
0.951897
My favorite example of coupling Value_Locate and Histogram is in the case of sparse data. For example, let's say we want to know which values are duplicated in the following data.
data = [5, 1000000000000ULL, 1000000000000ULL, 6]
The obvious answer is Histogram.
IDL> h = Histogram(data, OMIN=mindata) IDL> Print, Where(h gt 1) + mindata
But this will fail miserably because the required histogram has almost one trillion elements and would require almost 4TB of memory! That's ridiculous overkill given that there are only 3 distinct data values.
The solution is to use Value_Locate to map those values onto the set of integers from 0 to 2, and the run histogram on those mapped values. First we need to get a list of all the possible values that data can take on:
IDL> sorteddata = data[Sort(data)]
IDL> dataenum = sorteddata[Uniq(sorteddata)] IDL> Print, dataenum
5 6 1000000000000
Now we use the dataenum variable to map the original data to the set of integers.
IDL> mappeddata = Value_Locate(dataenum, data) IDL> Print, mappeddata 0 2 2 1
Then we run histogram.
IDL> h = histogram(mappeddata, min=0) IDL> print, h 1 1 2
And figure out which elements have more than one drop in a histogram bucket.
IDL> Print, dataenum[Where(h gt 1)] 1000000000000
This technique can be used to compress any sparse data set into a range that Histogram can run on. Any algorithmic tricks that are based on Reverse_Indices (and there are a great many!) can now be extended to work on sparse data sets.
![]()
Version of IDL used to prepare this article: IDL 7.0.3.
![]()
Copyright © 2009 David W. Fanning and Jeremy Bailin
Last Updated 24 June 2009
